-
从实验室到临床应用,AI抗体离我们有多远?
近年来,基于计算/人工智能 (AI)的抗体开发方法(简称AI抗体)成为热门话题。AI抗体技术的问世引发了资本市场的广泛关注,其潜在的行业革新价值可能为产业发展带来新的机遇与变革。我们将在这篇文章中重点介绍下AI抗体方法的优势、局限性及其最新进展。
01 AI抗体工具的出现
自20世纪末以来,单克隆抗体 (mAb) 已成为癌症靶向治疗的重要工具[1]。常规的基于生物体的免疫系统获得抗体的实验方法成本高昂且耗时,局限性较多。早期AI抗体设计的计算方法,受限于有限的结构数据和计算能力,阻碍了可靠的抗体-抗原相互作用模型的开发[2],导致结合预测不准确,限制了实用性并需要大量的体外验证。由于计算和人工智能 (AI) 技术近年来的爆炸性进展,人们它能希望能克服其中的许多局限性,在抗体开发流程中起到重要作用。因此我们这里,重点关注的是以人工智能 (AI),尤其是以机器学习 (ML) 和深度学习 (DL)为模型特征AI抗体方法。
人工智能 (AI)通过改进对抗体-抗原结构、相互作用、结构动力学和分子稳定性的预测,来改变抗体设计。AI 驱动的抗体设计方法通过整合蛋白质数据库(PDB)等海量结构数据,并借助 AlphaFold 等先进工具,显著提升了计算机模拟的效率和准确性。随着计算能力和云平台的增强,这些进步使得快速模拟能够在一定程度上补充传统的实验方法,提高抗体的亲和力、特异性和治疗潜力。AI 模型分析复杂的数据集,准确地预测抗体序列、3D 结构、互补决定区 (CDR)、互补位、表位和抗原-抗体相互作用。这些创新志于简化抗体设计、优化和测试,减少了时间和成本,同时解决了与可开发性和稳定性相关的挑战。
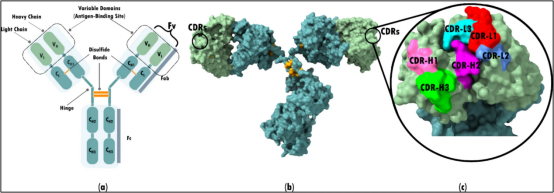
抗体的特异性结合能力取决于其六个高变区:CDR-H1、H2、H3(重链)以及CDR-L1、L2和L3(轻链)。人工智能在抗体设计和优化中, 尤其注重CDR开发、结构稳定性、折叠效率和CDR H3构象的预测,因为这是优化表位-互补位相互作用和增强疗效的关键因素。
02 最新AI抗体进展
AI方法通过降低成本和解决传统结构预测的局限性,补充了抗体发现、设计和优化中的实验方法。Rosetta、AlphaFold2 和 AlphaFold3、DeepAb、ABlooper和 DeepH3等工具促进了蛋白质和抗体结构预测的进步,支持在抗体设计和优化中使用结构数据。这些计算工具利用AI 和 ML 来加深对蛋白质折叠和相互作用的理解,有助于预测具有高亲和力潜力的抗体结构。
最新AI抗体模型结合高通量测序技术、日益丰富的结构数据库以及计算方法的优化,显著提升了抗体-抗原相互作用预测的准确性。在抗体设计领域,ML 和 DL 展现出强大潜力,特别是利用CNN、RNN等技术进行基于结构的蛋白质与抗体设计时表现突出。
在开发的工具中,很大部分是基于 Transformer 的模型。注意力机制的应用显著提升了SMILES序列预测、多任务学习以及化学分子和抗体结构设计与优化中的序列数据处理能力。例如:比较成功的有AlphaPanda,在一个案例研究中,识别高亲和力抗体方面取得了 87% 的成功率;AB-Gen,被用于设计 HER2 靶向抗体库,并通过模拟验证了关键残基;AntiBERTa,在互补位预测方面表现优异,并支持结构预测、人源化和 BCR 分析。其它工具还有 ESM-1v 、mCSM-AB2 等,预测突变对抗体-抗原结合亲和力等方面有着出色的准确度。
另一大阵营基于自然语言处理NLP模型,比如ProtT5、Transformer-XL和 BERT等,预训练语言模型在蛋白质工程、药物发现和功能注释方面具有潜力。AbBERT、ReprogBERT、AbImmPred、 AntiBERTy等,都在预测中有良好的表现。
其他的如 ProtGPT2、AbGPT、IgLM、AB-Gen、AntiBARTy Diffusion和 pAbT5,都在进一步推进抗体设计和工程,整合AI驱动的序列和结构优化,解决免疫学难题以及增强蛋白质工程,简化了方案。
不少工具如IgFold、ImmuneBuilder、DeepH3在 CDR 预测、生成和建模中的准确性都有所提高。下表是各种工具以及各自偏重的功能总结[3]。
已开发的工具及其应用
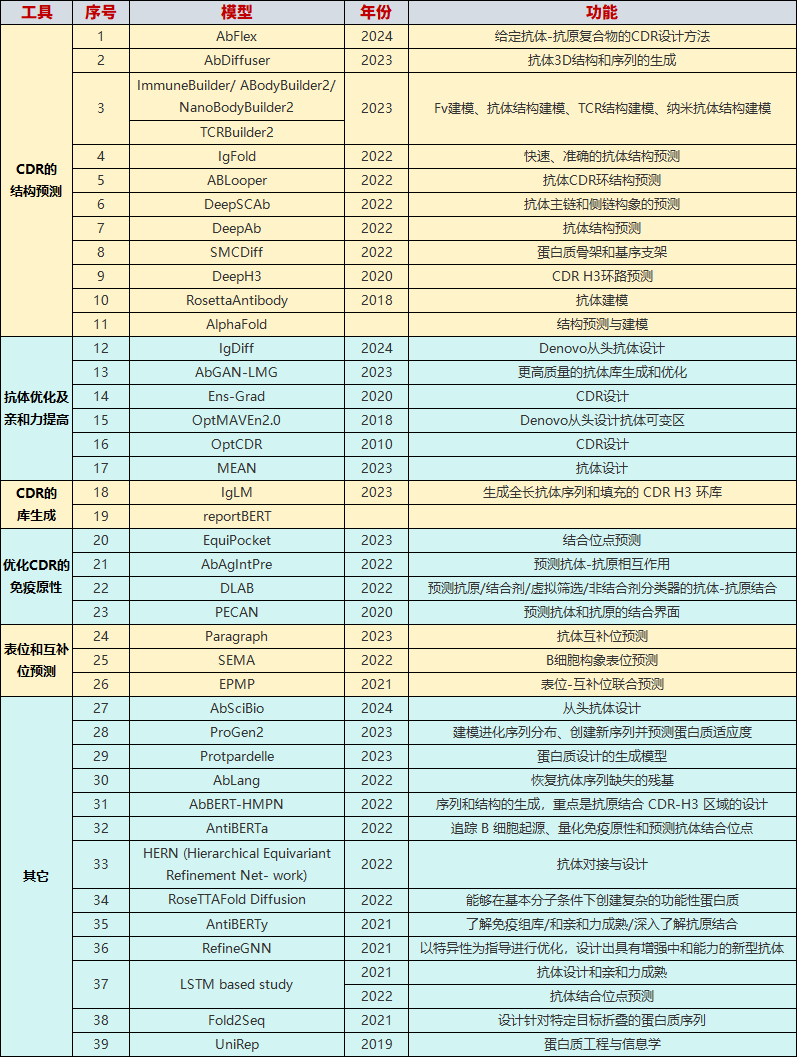
03 已开发的工具及其应用
尽管不断进步,抗体设计仍然面临挑战,包括预测限制、计算复杂性、数据稀缺等。许多关键的物理化学性质,如稳定性、溶解度、免疫原性和亲和力,很难仅从序列中预测,通常需要实验验证才能确认。训练方法和数据可用性的持续改进有可能增强其预测能力和适用性。AI模型可能面临泛化的限制,有时会产生已知抗体的变体,而不是完全新颖的候选抗体。过度拟合也是 AI 驱动的抗体设计中的一个挑战。由于生物系统的复杂性,AI 生成的抗体验证需要进行广泛的体外和体内测试。数据质量限制、模型可解释性、高计算成本以及抗体序列空间的巨大复杂性使得AI抗体设计在泛化能力、实际适用性以及对高质量实验数据的依赖方面均面临挑战。随着深度学习以及物理建模和实验验证的不断进步,AI抗体方法会在加速抗体开发方面发挥越来越重要的作用。
[1] Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7.
[2] Hummer AM, Abanades B, Deane CM. Advances in computational structure-based antibody design. Curr Opin Struct Biol. 2022;74:102379.
[3] Dewaker, V., Morya, V.K., Kim, Y.H. et al. Revolutionizing oncology: the role of Artificial Intelligence (AI) as an antibody design, and optimization tools. Biomark Res 13, 52 (2025).
-
磷酸化抗体的WB操作注意事项

-
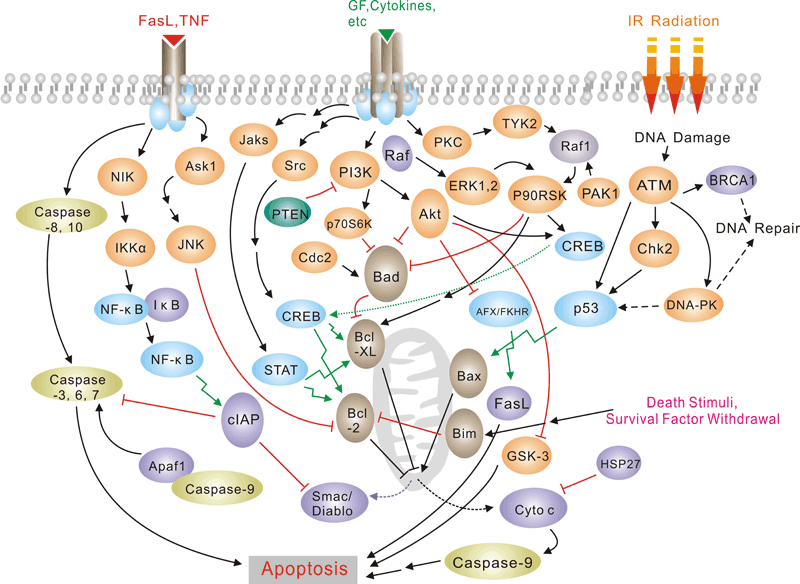
癌症/细胞凋亡信号通路
Pathway description:
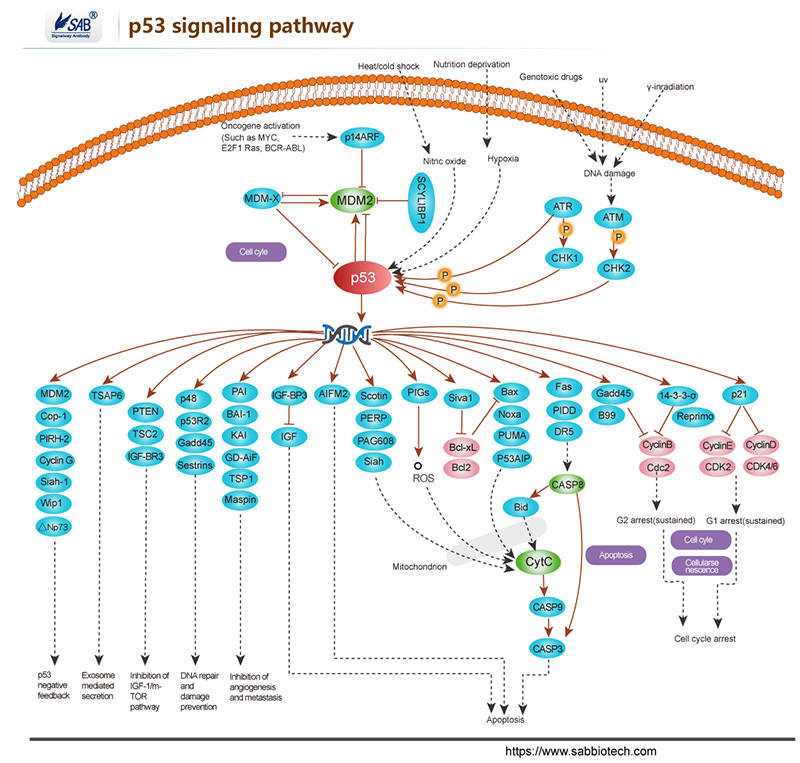
Apoptosis is a naturally occurring process by which a cell is directed to Programmed Cell Death. Apoptosis is based on a genetic program that is an indispensable part of the development and function of an organism and is a regulated physiological process leading to cell death. In apoptosis pathways, signaling results in the activation of a family of Cysteine Proteases, named Caspases that act in a proteolytic cascade to dismantle and remove the dying cell,are central regulators of apoptosis.Initiator caspases (including 8, 9,10) are closely coupled to pro-apototic signals. Once activated, these caspases cleave and activate downstream effector caspases (3,6,7) which in turn cleave cytoskeletal and nuclear proteins and induce apoptosis. Cytochrome C released from damaged mitochondria is coupled to the activation of caspase 9.
Pro-apoptotic stimuli include the FasL, TNF, DNA damage. Fas and TNFR activate caspases 8 and 10; DNA damage leads to the activation of caspase 9. Anti-apoptotic ligands including growth factors and cytokines activate AKT and p90RSK, which inhibit Bad and prevent cytochrome C release. TNFR can also stimulate an anti-apoptotic pathway by inducing IAP, which directly inhibits caspases 3, 7 and 9.
Alternatively, apoptosis is inhibited via an adaptor protein complex which activates NF-kB and induces survival genes including IAP. The Bcl-2 family of proteins regulate apoptosis by controlling mitochondrial permeability and the release of cytochrome C. The anti-apoptotic proteins Bcl-2 and Bcl-xL reside in the outer mitochondrial wall and inhibit cytochrome C release. The pro- apoptotic Bcl-2 proteins Bad, Bid, Bax and Bim reside in the cytosol but translocate to mitochondria following death signaling, where they promote the release of cytochrome C. Bad translocates to mitochondria and forms a pro-apoptotic complex with Bcl-xL. This translocation is inhibited by survival factors that induce the phosphorylation of Bad, leading to its cytosolic sequestration. Bax and Bim translocate to mitochondria in response to death stimuli, including survival factor withdrawal. Bcl-xL, Bcl-2 and Bax apparently influence the voltage-dependent anion channel (VDAC), which can control cytochrome C release. p53, activated following DNA damage, induces the transcription of Bax. Released cytochrome C binds Apaf1 and forms an activation complex with caspase9.
Selected Reviews:
Czerski L,Nune G..(2004)Apoptosome formation and Csapase activation:is it different in the heart.J Mol Cardiol.37(13),643-652.
Debatin KM, Krammer PH (2004) Death receptors in chemotherapy and cancer. Oncogene 23(16), 2950–66.
-
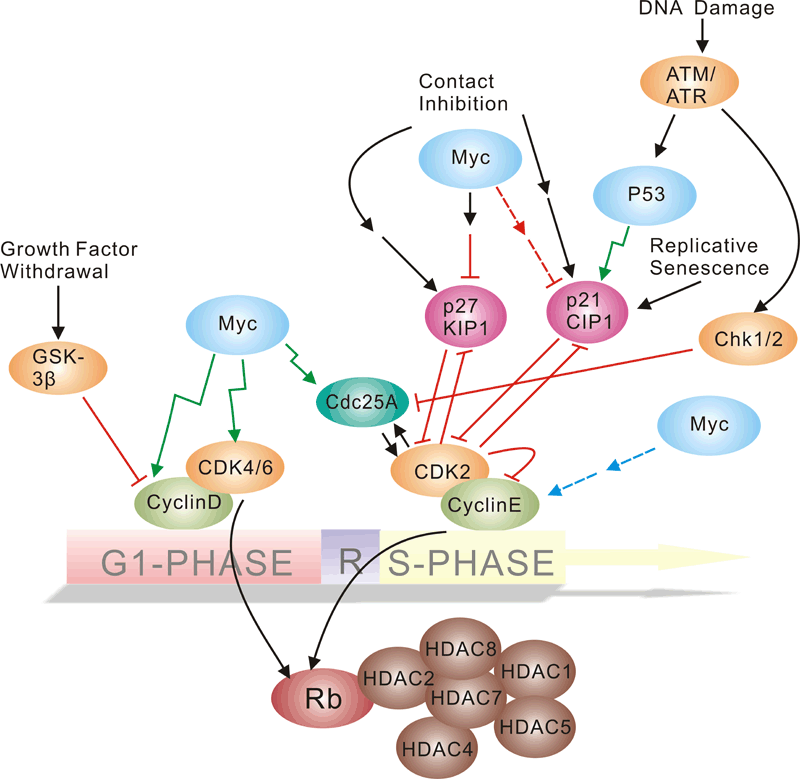
细胞周期信号通路
Pathway description:
Cell-division control affects many aspects of development. Caenorhabditis elegans cell-cycle genes have been identified over the past decade, including at least two distinct Cyclin-Dependent Kinases (CDKs), their cyclin partners, positive and negative regulators, and downstream targets. The balance between CDK activation and inactivation determines whether cells proceed through G1 into S phase, and from G2 to M, through regulatory mechanisms that are conserved in more complex eukaryotes. Many different stimuli exert checkpoint control including TGF, DNA damage, contact inhibition, replicative senescence, and growth factor withdrawal. G1 phase CDKs and their inhibitors (CKIs) are central to the pathways that regulate commitment to cellular division in response to positive as well as negative growth effectors. Many checkpoints are deregulated in oncogenesis, and this is often due to alterations in cyclin-CDK complexes.CDK activity is modulated by cyclin binding, phosphorylation, and CKIs, including the INK4 proteins and the closely related inhibitors p21Cip1 and p27Kip1. The downstream targets of CDKs and their modulation by TGF-beta and other growth factors include proteins of the retinoblastoma family, and the related modulation of the transcriptional activity of the E2F family members.
Selected Reviews:
Dyson, N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262.
Ravitz MJ, Wenner CE. (1997)Cyclin-dependent kinase regulation during G1 phase and cell cycle regulation by TGF-beta. Adv Cancer Res. 71:165-207.
van den Heuvel S.(2005) Cell-cycle regulation. WormBook. 21:1-16.
-
染色质/转录信号通路
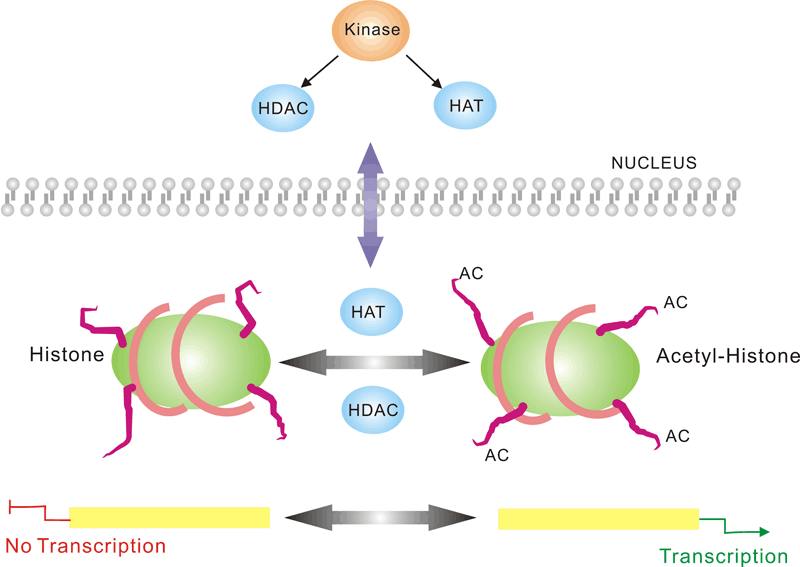
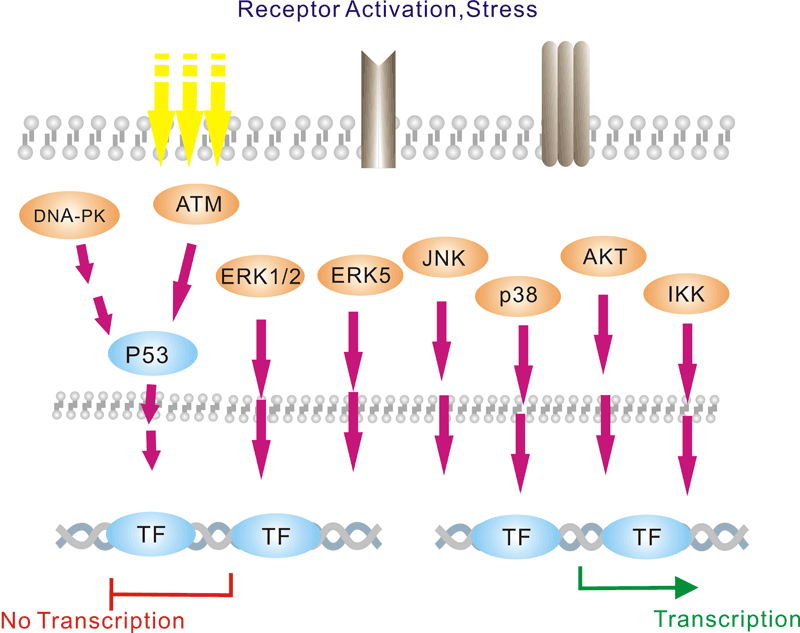
Pathway description:
Chromatin is the substance which becomes visible chromosomes during cell division. Its basic unit is nucleosome, composed of 146 bp DNA and eight histone proteins. The structure of chromatin is dynamically changing, at least in part, depending on the need of transcription. In the metaphase of cell division, the chromatin is condensed into the visible chromosome.
Transcription is the process through which a DNA sequence is enzymatically copied by an RNA polymerase to produce a complementary RNA. In eukaryotes, it takes place in the nucleus, mitochondria and chloroplast.
Protein acetylation plays a crucial role in regulating chromatin structure and transcriptional activity. Acetylation complexes or deacetylation complexes can be recruited to DNA-bound transcription factors (TFs) in response to signaling pathways. Histone hyperacetylation by histone acetyltransferases (HATs) is associated with transcriptional activation, presumably by remodeling nucleosomal structure into an open conformation more accessible to transcription complexes. Conversely, histone deacetylation by deacetylation complexes (such as HDAC) is associated with transcriptional repression reversing the chromatin remodeling process.Several transcriptional coactivators and corepressors possess intrinsic acetylase or deacetylase enzymatic activities, respectively. Site-specific acetylation of a growing list of non-histone proteins, including p53 and E2F, has been shown to play an important role in transcriptional regulation and cell proliferation.
Selected Reviews:
Davie JR, Samuel SK, Spencer VA, et al. (1999)Organization of chromatin in cancer cells: role of signalling pathways. Biochem Cell Biol.77(4):265-75.Spencer VA, Davie JR.(2000)Signal transduction pathways and chromatin structure in cancer cells. J Cell Biochem Suppl. 35:27-35.
Crosio C, Heitz E, Allis CD,et al. (2003)Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 116(Pt 24):4905-14.
-
细胞骨架/粘连信号通路
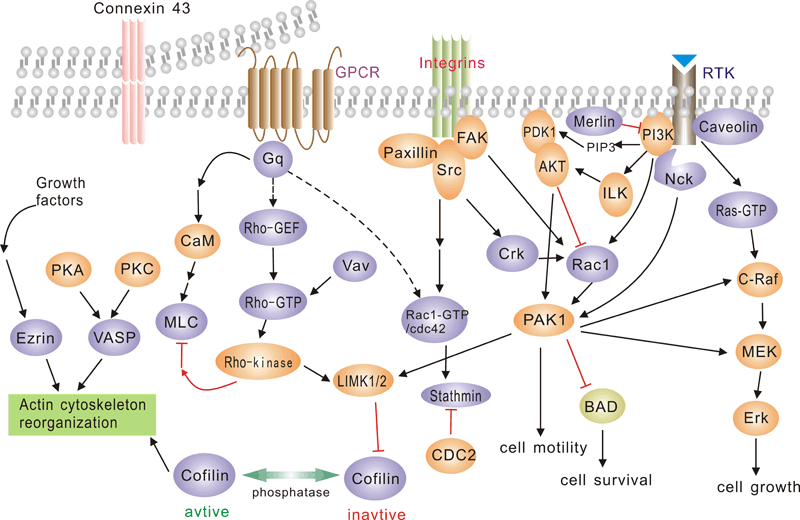
Pathway description:
G protein-coupled receptors (GPCRs), comprise a large protein family of transmembrane receptors that sense molecules outside the cell and activate inside signal transduction pathways and, ultimately, cellular responses. GPCRs are activated by an external signal in the form of a ligand or other signal mediator. This creates a conformational change in the receptor, causing activation of a G protein.The extracellular parts of the receptor can be glycosylated. These extracellular loops also contain two highly-conserved cysteine residues that form disulfide bonds to stabilize the receptor structure.
The family of p21-activated protein kinases (PAKs) is composed of serine-threonine kinases whose activity is regulated by the small guanosine triphosphatases (GTPases), Rac and Cdc42 In mammalian cells, PAKs have been implicated in the regulation of mitogen-activated protein cascades, cellular morphological and cytoskeletal changes, neurite outgrowth, and cell apoptosis.
Focal adhesion kinase (FAK) is a nonreceptor protein tyrosine kinase involved in integrin-mediated control of cell behavior. Following cell adhesion to components of the extracellular matrix, FAK becomes phosphorylated at multiple sites, including tyrosines 397, 576, and 577. Tyr-397 is an autophosphorylation site that promotes interaction with c-Src or Fyn. Tyr-576 and Tyr-577 lie in the putative activation loop of the kinase domain, and FAK catalytic activity may be elevated through phosphorylation of these residues by associated Src family kinase. The clustering of integrins at these sites attracts a large complex of proteins and initiates intracellular regulatory processes, by which such events as cell migration and anchorage-dependent differentiation are controlled. FAK is a protein tyrosine kinase which is recruited at an early stage to focal adhesions and which mediates many of the downstream responses. FAK subsequently interacts with a number of down-stream signalling proteins, including the adaptor protein Grb2 and the p85 _-subunit of phosphatidylinositol 3 kinase (PI3 kinase)
References:
Daniels, R. H. & Bokoch, G. M. (1999) p21-Activated protein kinase: a crucial component of morphological signaling? Trends Biochem. Sci. 24, 350-355.
Lei, M. et al. (2000) Structure of PAK1 in an Autoinhibited Conformation Reveals a Multistage Activation Switch . Cell 102, 387-397
Joost P, Methner A(2002) Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol 3 (11), research0063.1-0063.16.
Bjarnadottir TK. et al. (2006) Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 88 (3), 263-73.
Astier A. et al. (1997) The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after beta1-integrin stimulation in B cells and binds to p130cas. J Biol Chem.272,228–232.
Avraham S,.et al. (1995) Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J Biol Chem. 270,27742–27751.
-
DNA损伤/修复信号通路
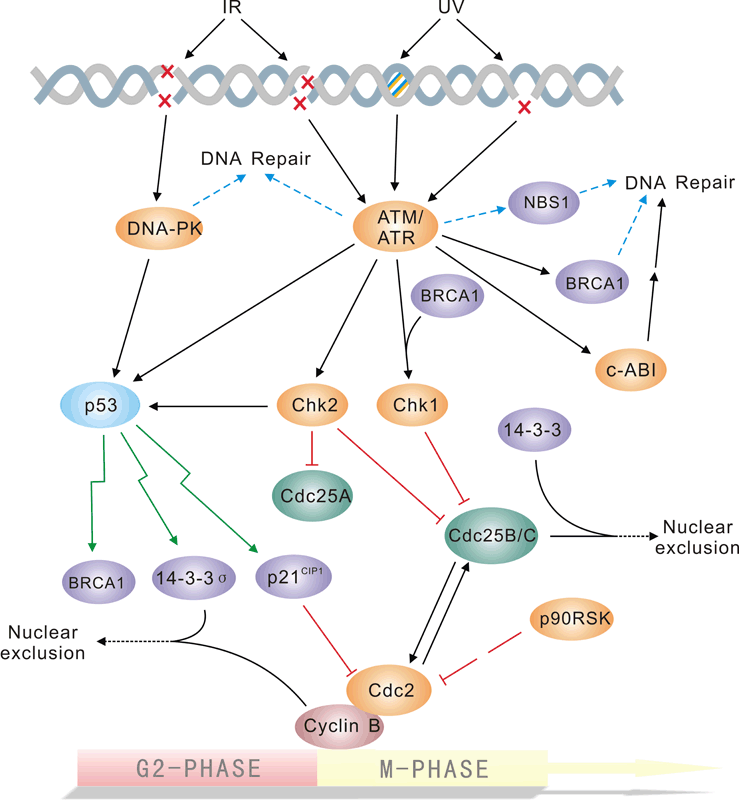
Pathway description:
The repair of DNA lesions that occur endogenously or in response to diverse genotoxic stresses is indispensable for genome integrity. DNA lesions activate checkpoint pathways that regulate specific DNA-repair mechanisms in the different phases of the cell cycle. Checkpoint- arrested cells resume cell-cycle progression once damage has been repaired, whereas cells with unrepairable DNA lesions undergo permanent cell-cycle arrest or apoptosis. The G2/M DNA damage checkpoint prevents the cell from entering mitosis (M phase) if the genome is damaged. Environmental DNA-damaging agents include UV light and ionizing radiation. Many of the checkpoint proteins are activated in response to DNA damaged, and a number of phosphorylation events in DNA repair mechanism depend on activation of these kinases. The Mec1/Rad3/ATM/ATR family act early in the checkpoint pathways either as DNA damage detectors or in close association with such detectors. The tumor suppressor P53 is central to the higher eukaryotic checkpoint controls in DNA damage-induced G1 arrest through transcriptional induction of the cyclin-dependent kinases Chk1 and Chk2, and inhibitor p21.Activation of p53 in response to DNA damage and other stresses involves complex posttranslational modification of p53 and its negative modulater MDM2,which is itself induced by p53 at the transcriptional level.
Selected Reviews:
Norbury CJ,Hickson ID.(2001)Cellular responses to DNA damage.Annu Rev Pharmacol Toxicol.41:367-461.
Bartek J,Lukas J.(2003)Chk1 and Chk2 kinases in checkpoint control and cancer.Cancer Cell.3(5)421-9.
Branzei D, Foiani M. (2008)Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 9(4):297-308.
-
胰岛素/葡萄糖代谢信号通路
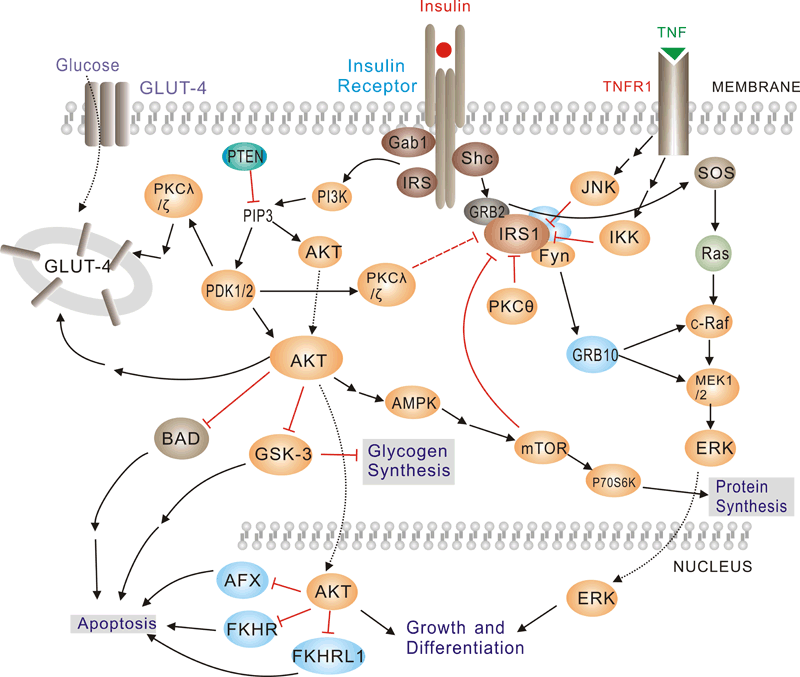
Pathway description:
Insulin has diverse effects on cells including stimulation of glucose transport, gene expression and alterations of cell morphology. Insulin signaling begins with either the activation or substrate kinase activity of the insulin receptor (IR), which is the only component of the pathway that is unique to insulin action. The insulin receptor belongs to a subfamily of receptor tyrosine kinases. Activation of the IR can be impaired by post-translational modifications of the protein involving serine phosphorylation, or by binding to inhibiting proteins such as PC-1 or members of the SOCS or Grb protein families. Insulin elicits a diverse array of biological responses by binding to its specific receptor. The hormone mediates these effects by activation of signaling pathways which adaptor molecules such the IRS, the SHC and the GRB2 lipid kinases such as PI3K serine, threonine and tyrosine.
Selected Reviews:
ALAN R,SALTIEL,et al.(2001)Insulin signaling and the regulation of glucose and lipid metabolism.Nature.414(6865):799-806.
Ogawa W,Matozaki T,et al.(1998)Role of binding proteins to IRS-1 in insulin signaling. Mol Cell Biochem.182(1-2):13-22.
Youngren JF. (2007)Regulation of insulin receptor function. Cell Mol Life Sci. 64(7-8):873-91
Johnston AM, Pirola L, Van Obberghen E.2003Molecular mechanisms of insulin receptor substrate protein-mediated modulation of insulin signalling. FEBS Lett. 546(1):32-6.
-
MAPK signaling pathway
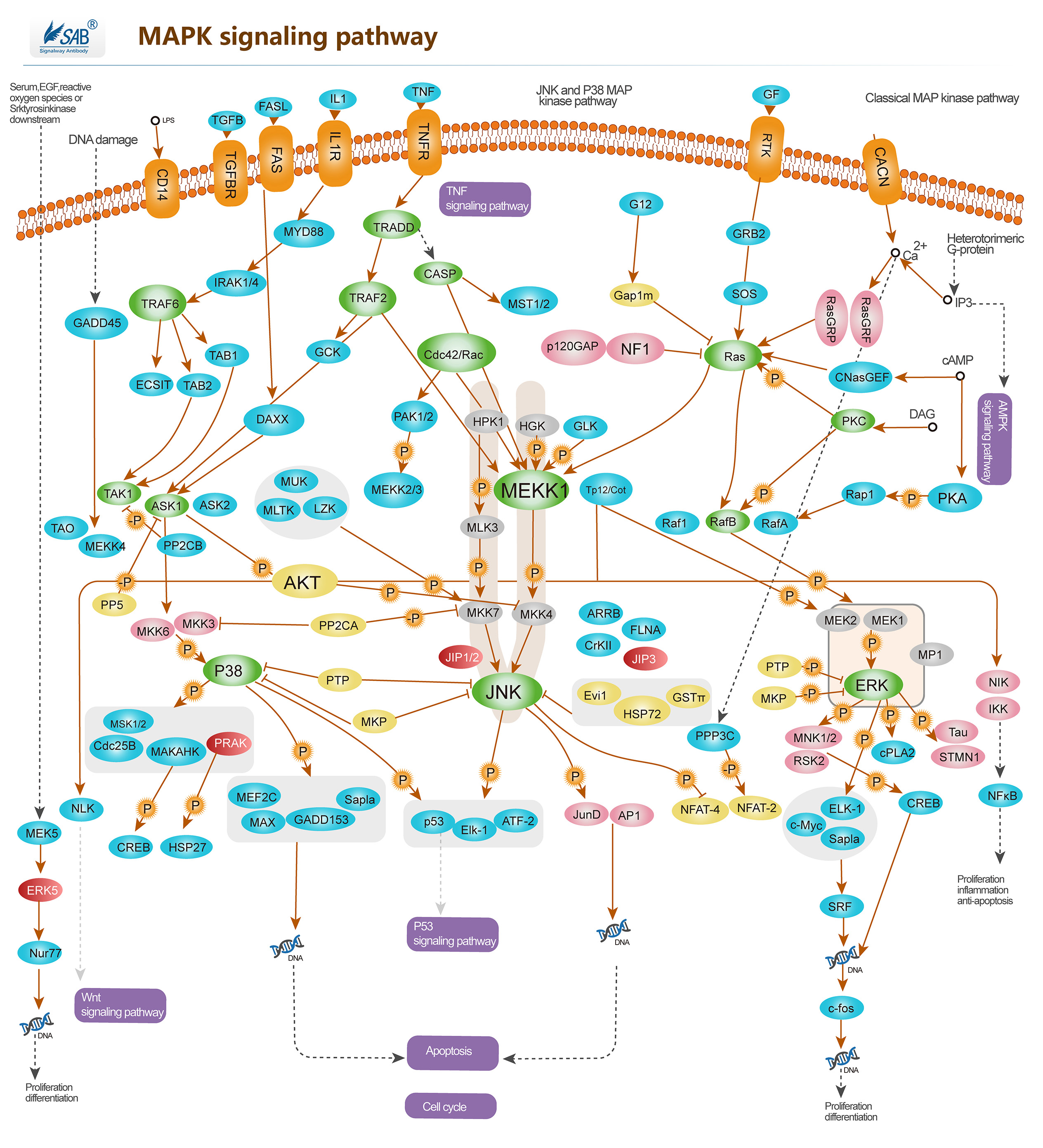
Pathway description:
Intracellular signaling cascades are the main routes of communication between the Plasma membrane and regulatory targets in various intracellular compartments. Sequential activation of Kinases is a common mechanism of signal transduction in many cellular processes. During the past decade, several related intracellular signaling cascades have been elucidated, which are collectively known as MAPK (Mitogen-Activated Protein Kinase) signaling cascades. The MAPKs are a group of protein Serine/threonine Kinases that are activated in response to a variety of extracellular stimuli and mediate signal transduction from the cell surface to the nucleus. In combination with several other signaling pathways, they can differentially alter phosphorylation status of numerous proteins. There are four major groups of MAPKs in mammalian cells-the ERKs, the p38MAPKs, the JNKs and the ERK5 or BMK cascades. These MAPKs are actived by dual phosphorylation at the tripeptide motif Thr-Xaa-Tyr. Each MAPK pathway contains a three-tiered kinase cascade comprising a MAPKKK, a MAPKK and the MAPK. Frequently, a MAPKKKK activates the MAPKKK.The MAPKKK then phosphorylates a dual-specificity protein kinase MAPKK, which in turn phosphorylates the MAPK.p38MAPKs are members of the MAPK family that are activated by a variety of environmental stresses and inflammatory cytokines. Stress signals are delivered to this cascade by members of small GTPases of the Rho family (Rac, Rho, Cdc42). As with other MAPK cascades, the membrane-proximal component is a MAPKKK, typically a MEKK or a mixed lineage kinase (MLK). The MAPKKK phosphorylates and activates MKK3/6, the p38 MAPK kinase. MKK3/6 can also be activated directly by ASK1, which is stimulated by apoptotic stimuli. p38 MAPK is involved in regulation of Hsp27 and MAPKAP-2 and several transcription factors including ATF2, STAT1, the Max/Myc complex, MEF-2, ELK-1 and indirectly CREB via activation of MSK1.
Selected Reviews:Wada T,Penninger JM.(2004)Mitogen-activated protein kinase in apoptosis regulation. Oncogene. 23(16),2838
Panka DJ,Atkins MB,et al.(2006)Targeting the mitogen-activated protein kinase pathway in the treatment of malignant melanoma.Clin Cancer Res.12(7Pt2),2371s.
Dunn KL, Espino PS, Drobic B, et al.(2005)The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 83(1):1-14
-
TGFb/smads signaling pathway
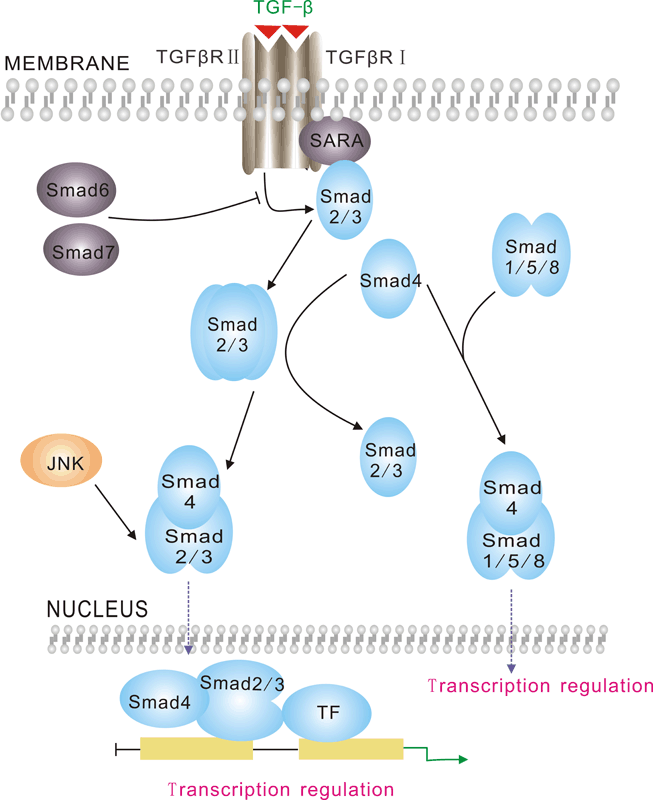
-
翻译信号通路
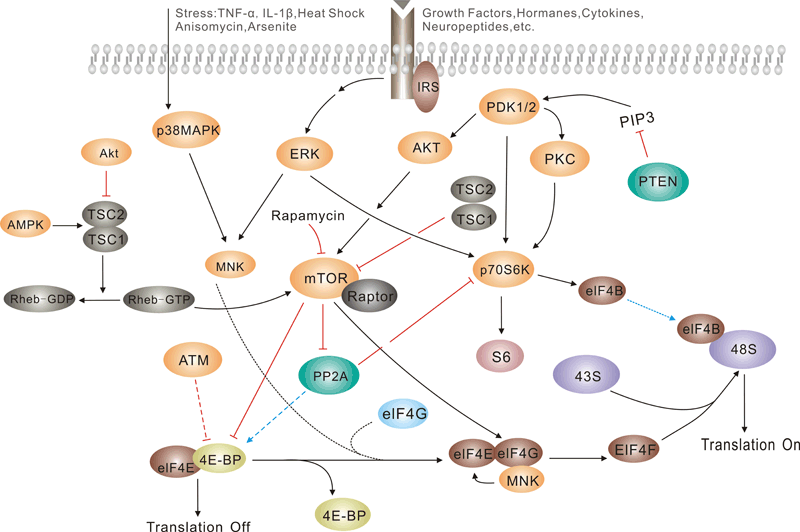
Pathway description:
Translation initiation is a complex process in which initiator tRNA, 40S, and 60S ribosomal subunits are assembled by eukaryotic initiation factors (eIFs) into an 80S ribosome at the initiation codon of mRNA. The cap-binding complex eIF4F and the factors eIF4A and eIF4B are required for binding of 43S complexes (comprising a 40S subunit, eIF2/GTP/Met-tRNAi and eIF3) to the 5′ end of capped mRNA but are not sufficient to promote ribosomal scanning to the initiation codon.
The eukaryotic translation initiation factor 4F (eIF4F) mediates 40S ribosomal subunit binding to the 5'end of capped mRNA. eIF4F is a complex containing three proteins: eIF4E, the cap-binding subunit; eIF4A, an RNA-dependent ATPase/ATP-dependent RNA helicase; and eIF4G, a high-molecular-weight protein that acts as a scaffold for binding eIF4E and eIF4A. In addition, eIF4G interacts with the 40S ribosome binding factor eIF3 and the poly(A)-binding protein, thereby establishing a critical link between mRNA and the ribosome. The various eIF4F subunits are expressed to remarkably different levels in most cell types, with the eIF4E subunit being the least abundant.
References:
Anthony, D. D., and W. C. Merrick. (1991) Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J. Biol. Chem. 266,10218-10226 Avdulov, S. et al. (2004) Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5,553-563.
Gingras, A. C.,et al. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68,913-963.
Hentze, M. W. (1997) eIF4G: a multipurpose ribosome adapter? Science 275,500-501
Hershey, J. W. B., and W. C. Merrick.(2000) Translational control of gene expression Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
p53 signaling pathway
-
Hippo signaling pathway

-
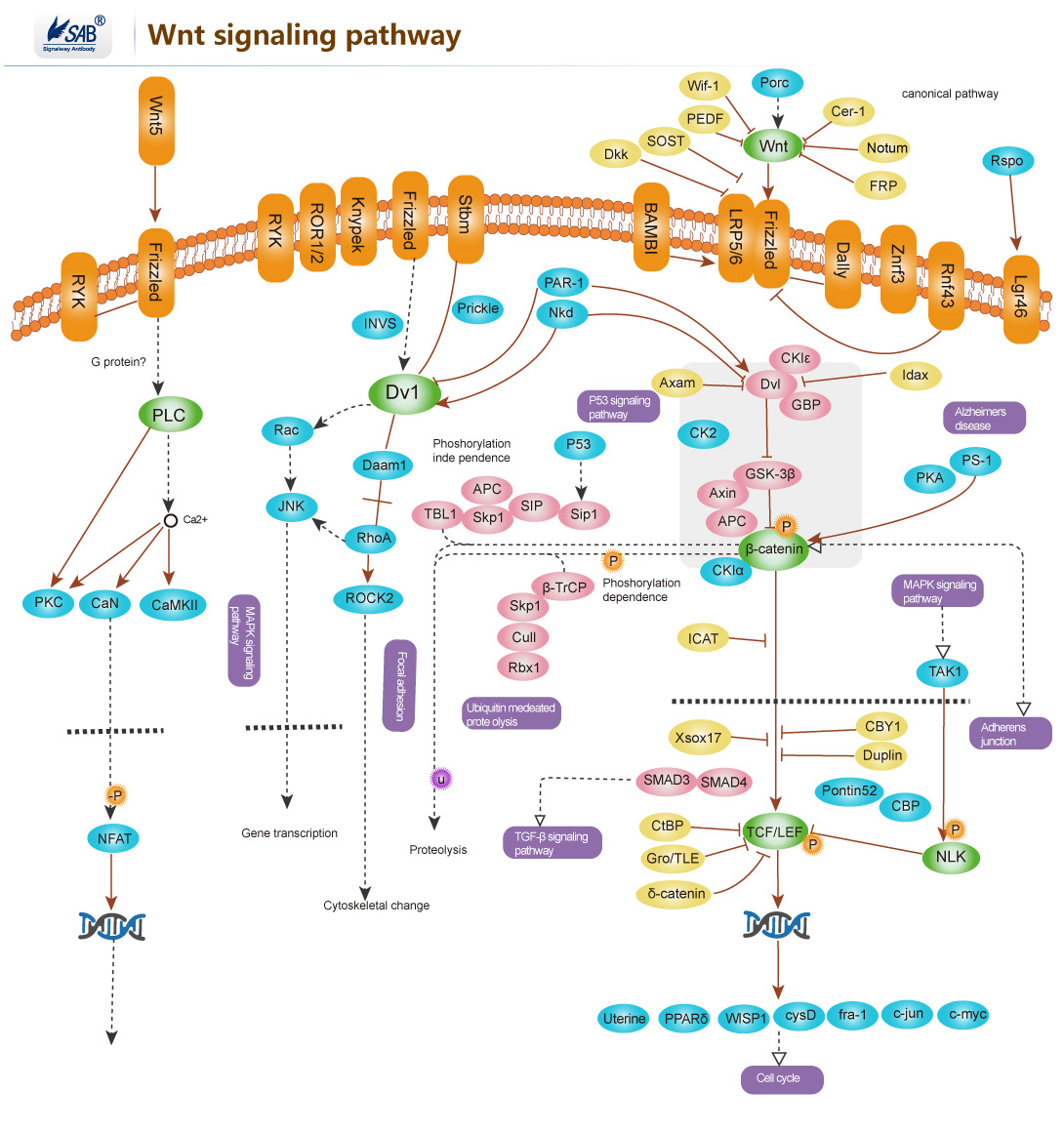
Wnt signaling pathway
-
ErbB signaling pathway
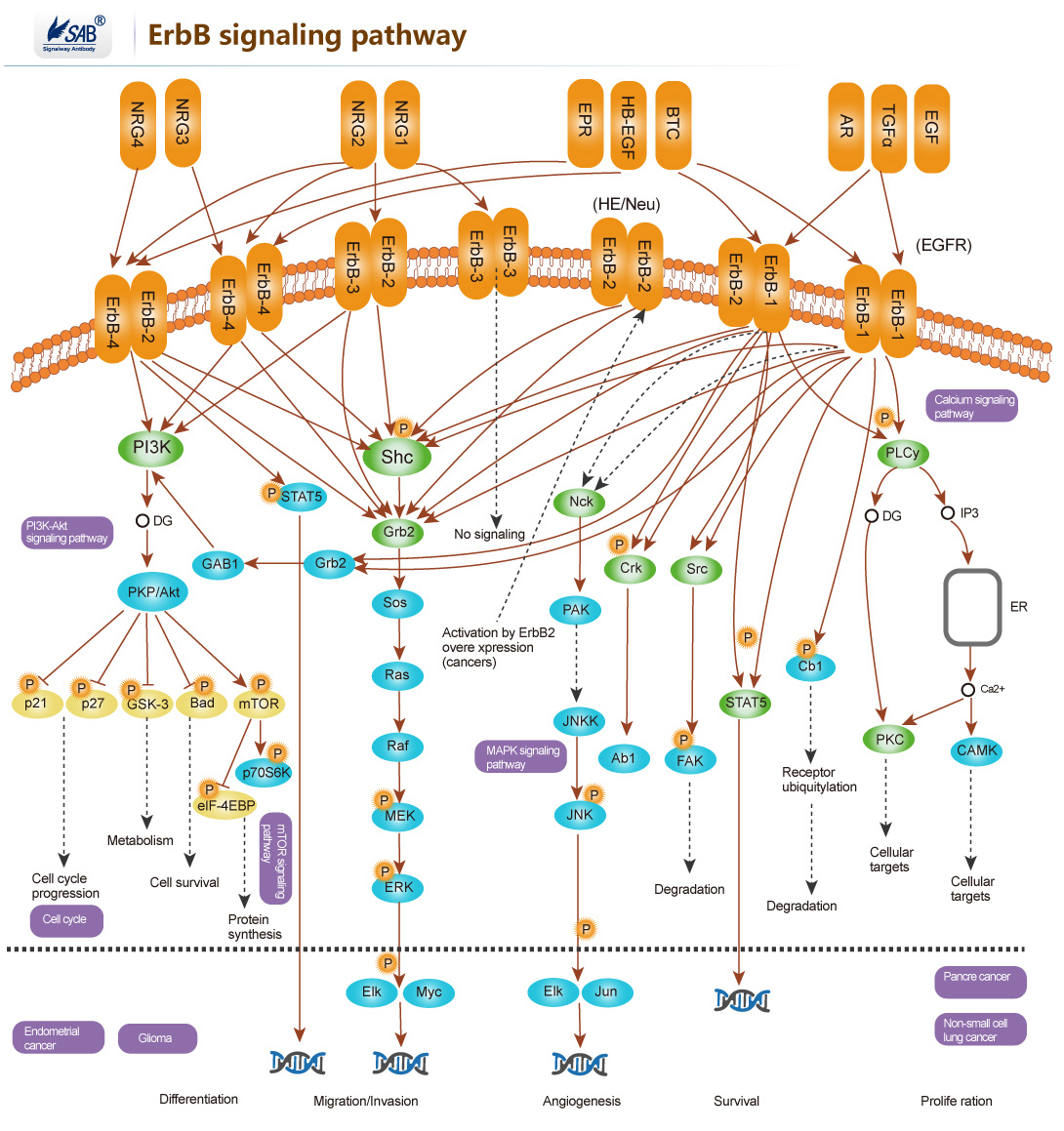
ERBB2编码的蛋白属于表皮生长因子受体家族,该家族包括HER1(erbB1,EGFR)、HER2(erbB2,NEU)、HER3(erbB3)及HER4(erbB4),也属于受体酪氨酸激酶家族。ERBB2没有与配体结合的结构域,但可以与家族成员形成异源二聚体来结合配体如表皮生长因子(EGF),从而通过磷酸化激活下游蛋白如MAPK和PI3K。ERBB2的过表达在多种肿瘤中被发现,常见的为乳腺癌和卵巢癌。
-
T-CELL Pathway
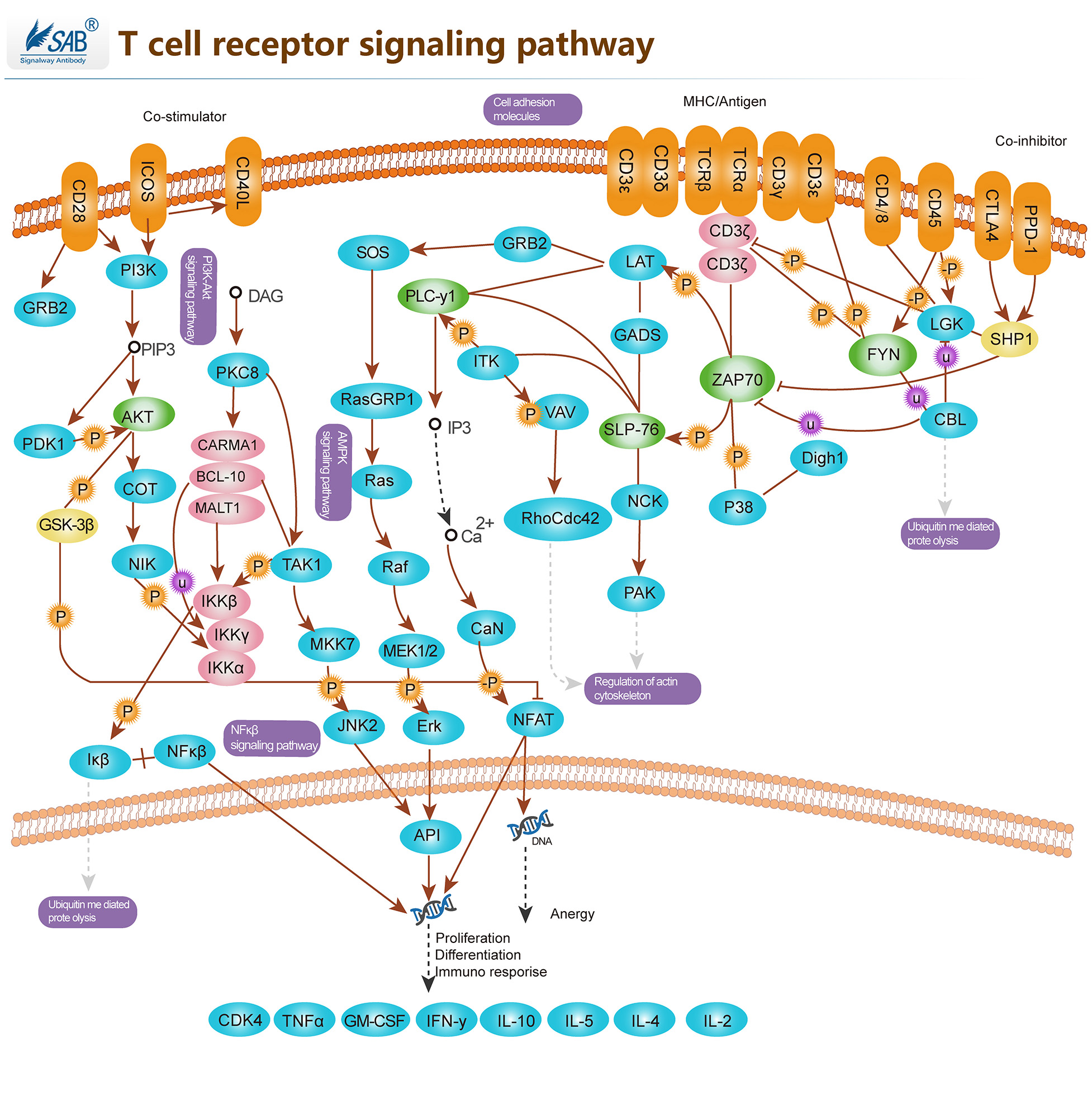
T细胞受体(TCR)在T细胞的功能和免疫突触的形成中起着关键作用。它在T细胞和抗原呈递细胞(APC)之间提供连接。TCRs激活促进了一系列信号级联,最终通过调节细胞因子的产生、细胞存活、增殖和分化来决定细胞的命运。T淋巴细胞的激活是免疫系统有效反应的关键事件。TCR激活受各种共刺激受体调节。CD28在T细胞激活过程中提供了一种必要的共刺激信号,可增加IL-2(白细胞介素-2)的产生,增加T细胞增殖,并防止无能和细胞死亡的诱导。通过B7-1或B7-2连接CD28有助于使T细胞和抗原呈递细胞膜接近。除CD28外,许多其他跨膜受体也调节TCR信号的特定元件,如CD45和CD4。TCR激活的早期事件是淋巴细胞蛋白酪氨酸激酶(Lck)使TCR/CD3复合物胞浆侧的免疫受体酪氨酸基激活基序(ITAM)磷酸化。CD45受体酪氨酸磷酸酶调节Lck和其他Src家族酪氨酸激酶的磷酸化和激活。TCR激活还通过激活ZAP70下游的GTP结合蛋白Rac和PAK导致细胞骨架重排。TCR信号的负调控也很重要,以检查与该通路相关的免疫反应过度激活。SIT(SHP2相互作用跨膜衔接蛋白)是最近发现的一种跨膜衔接蛋白,通过ITIM(免疫受体酪氨酸基抑制基序)与SHP2(含SH2的蛋白酪氨酸磷酸酶-2)相互作用,该复合物是TCR介导信号的关键负调节器。此外,CTLA4(细胞毒性T淋巴细胞抗原-4)也对T细胞活化产生负性调节。跨膜蛋白CTLA4也可作为天然抑制剂。一旦T细胞被激活,无论什么疾病过程使其启动,身体都有一个自然过程来关闭T细胞途径,使其不会失控。
-
mTOR Pathway
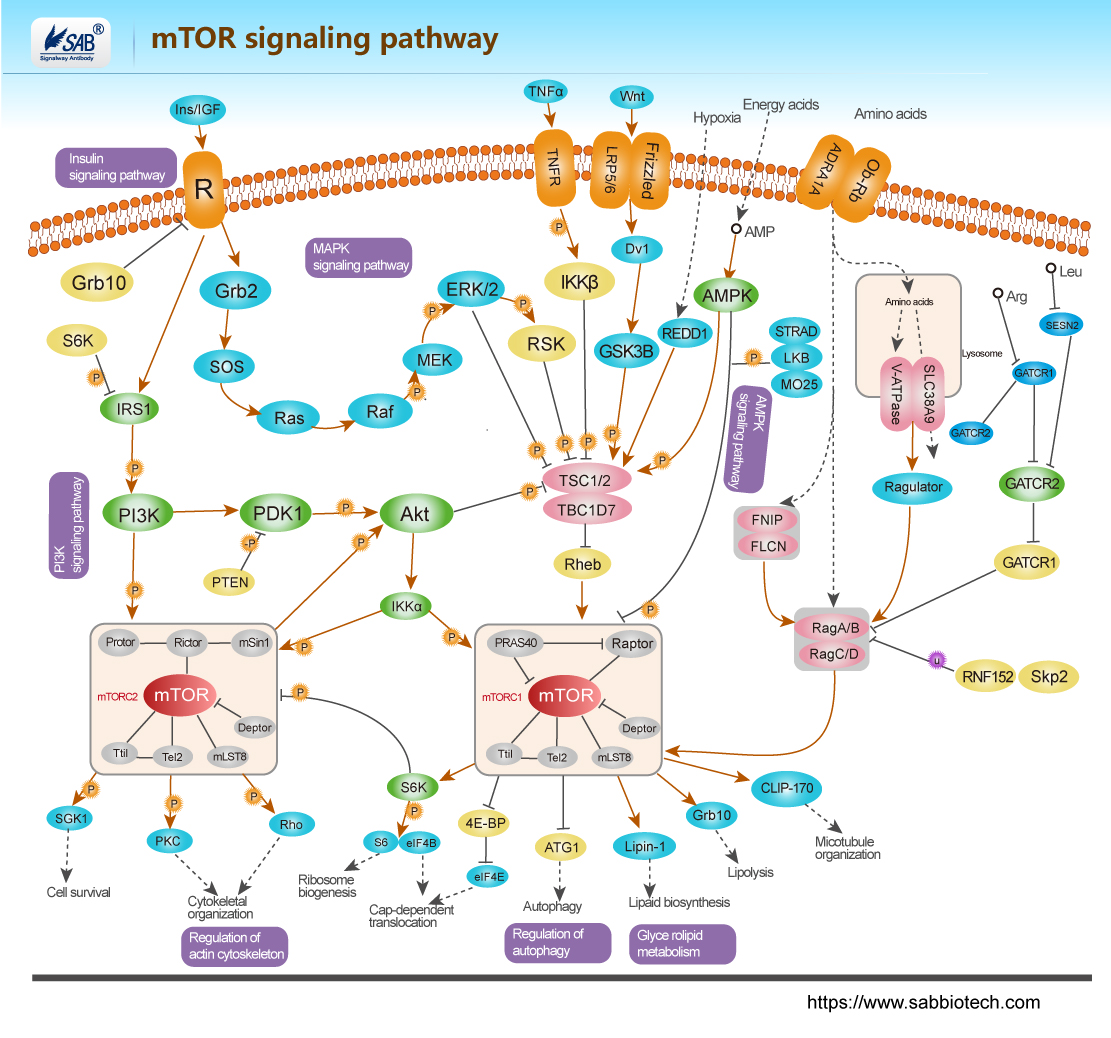
mTOR 通路受多种细胞信号的调控,包括有丝分裂生长因子、胰岛素等激素、营养素(氨基酸、葡萄糖)、细胞能量水平和应激条件。PI3K/Akt(v-Akt小鼠胸腺瘤病毒癌基因同源1)信号转导通路是通过mTOR 传递信号的主要通路,在介导细胞存活和增殖中起重要作用。通过 PI3K/Akt 通路的信号是由与细胞膜上的受体结合的生长因子的有丝分裂刺激启动的。这些受体包括IGFR (胰岛素样生长因子受体)、PDGFR (血小板衍生生长因子受体)、EGFR (表皮生长因子受体)和HER 家族。来自激活的受体的信号直接传递到PI3K/Akt 通路,或者,也可以通过由致癌蛋白RAS 激活的生长因子受体激活。RAS 是另一个信号转导的中枢开关,而且已证实是MAPK (丝裂原活化蛋白激酶)信号转导通路的关键激活子。胰岛素也可通过IRS1/2 (胰岛素受体底物-1/2)激活PI3K/Akt 通路。胰岛素结合激活IR (胰岛素受体)酪氨酸激酶,使IRS1 或IRS2 磷酸化。PI3K 通过P85 调节亚基中的SH2 (Src-Homology-2)结构域与磷酸化IR 结合。这种相互作用激活了p110 催化亚基。然后,PI3K 催化膜结合的PIP2 (磷脂酰肌醇(4,5)二磷酸)转化为PIP3 (磷脂酰肌醇(3,4,5)-三磷酸)。PIP3 然后与Akt 的pleckstrin 同源结构域结合,通过二聚化和暴露其催化位点而导致Akt 的激活。
-
JAK-STAT Pathway
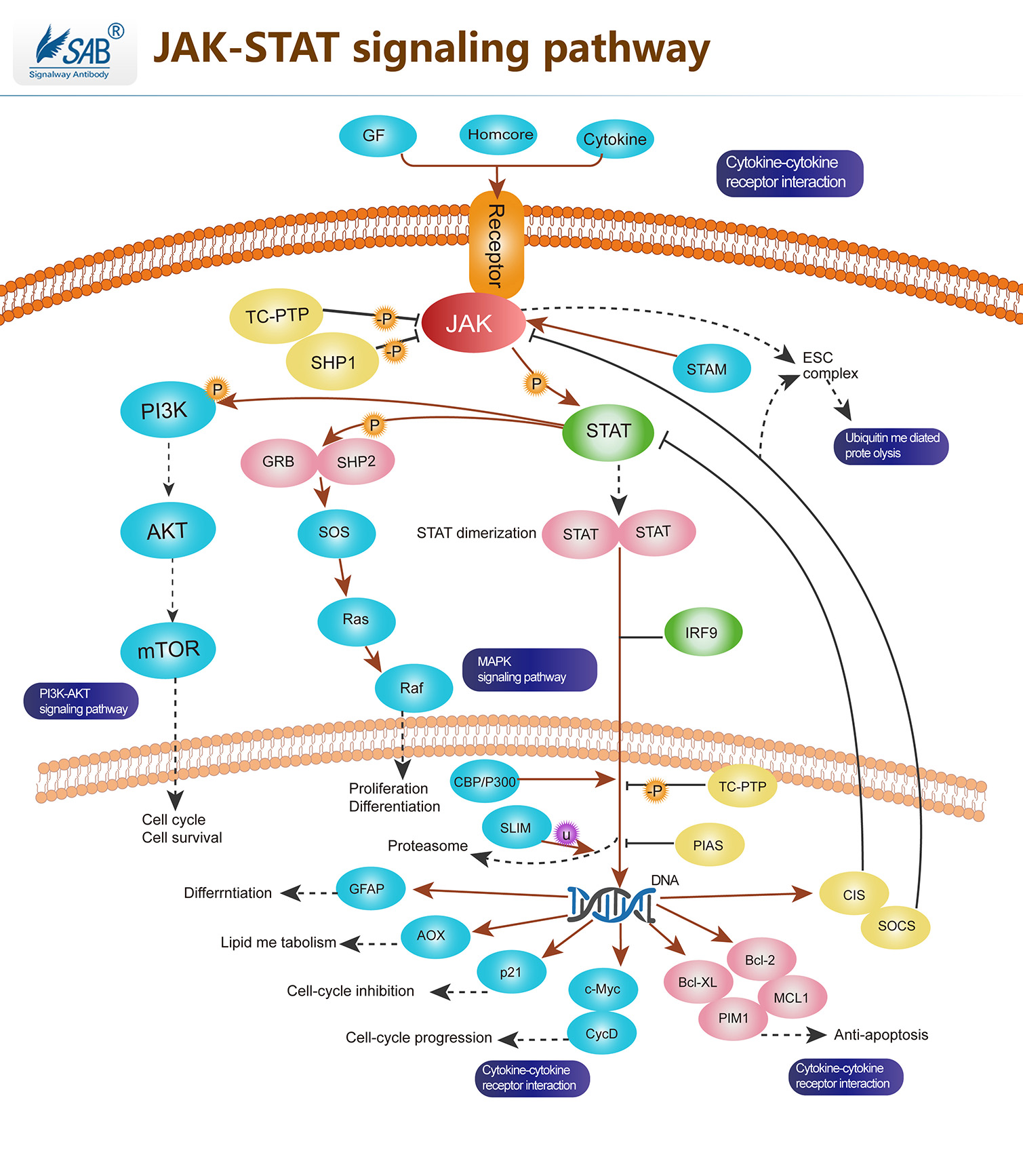
JAK -STAT途径是多种细胞因子和生长因子的主要信号传导机制。JAK激活刺激细胞增殖,分化,细胞迁移和凋亡。这些细胞事件对于造血,免疫发育,乳腺发育和泌乳,脂肪形成,两性性生长和其他过程至关重要。JAK(janus kinase)是一类非受体酪氨酸激酶家族,已发现四个成员,即Jak1 、Jak2 、Jak3 和Tyk2。JAK的N端结构域与受体相结合,C端为激酶结构域。每种激酶成员与特异的细胞因子受体结合。JAK的底物为STAT,即信号转导子和转录激活子(signal transducer and activator of transcription,STAT),N端具有SH2结构域和核定位信号(NLS),中间为DNA结合域,C端有保守的,对其活化至关重要的酪氨酸残基。共发现7个STAT家族成员,分别命名为STAT1至STAT7。STAT被JAK磷酸化后发生二聚化,然后穿过核膜进入核内调节相关基因的表达,这条信号通路称为JAK-STAT途径。
-
Autophagy Pathway
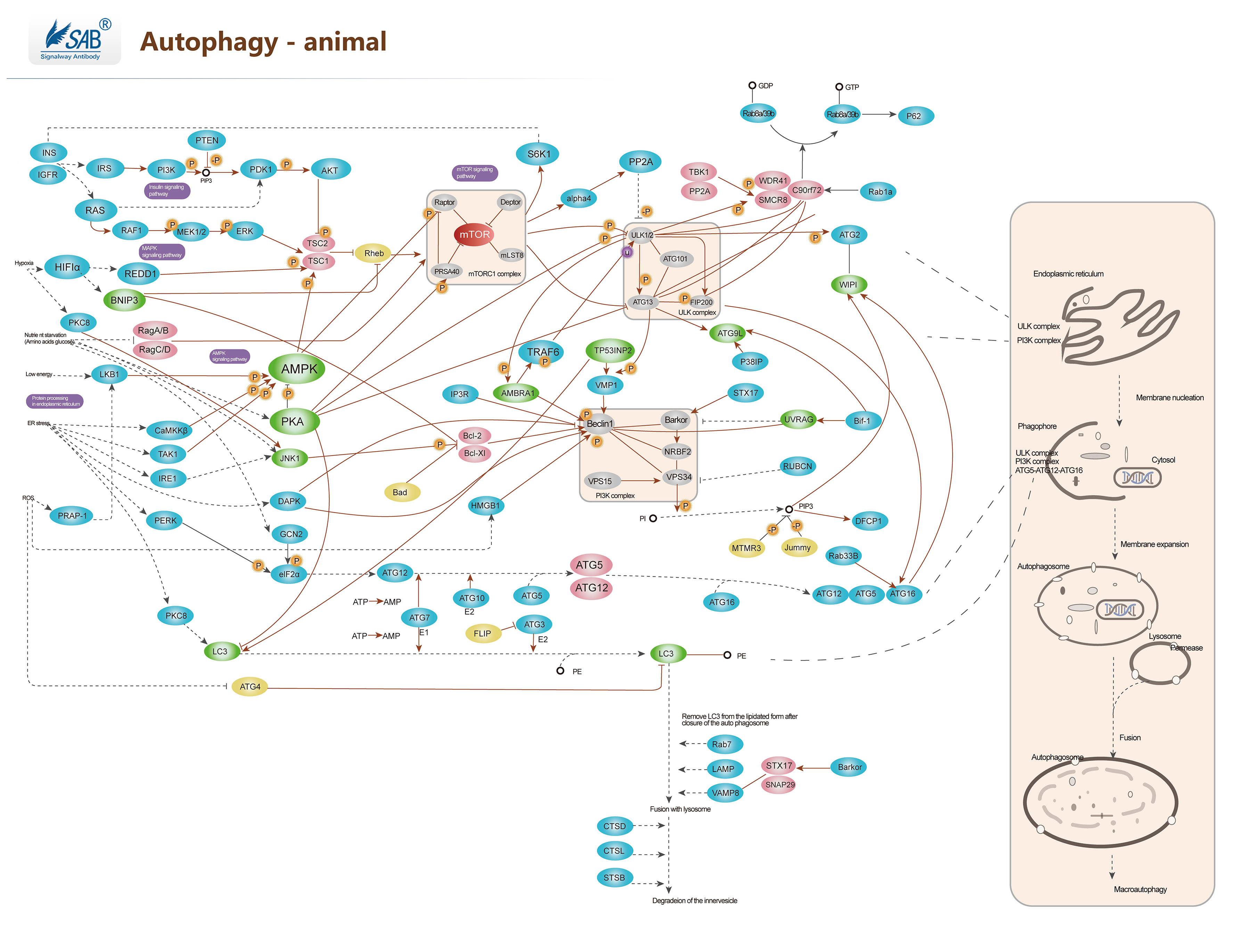
自噬 Autophagy,或称自体吞噬是一个涉及到细胞自身结构通过溶酶体机制,负责将受损的细胞器、错误折叠的蛋白及其他大分子物质等运送至溶酶体降解并再利用的进化保守过程。自噬是广泛存在于真核细胞的现象,并且可分为巨自噬、微自噬和分子伴侣介导的自噬三大类。这是一个受到紧密调控的步骤,此步骤是细胞生长、发育与稳态中的常规步骤,帮助细胞产物在合成、降解以及接下来的循环中保持一个平衡状态。目前已有多份研究表明自噬在许多细胞的分化进程中被不同程度地激活,例如参与血管生成、成骨分化、脂肪生成、神经发生等过程。自噬效应的发生取决于自噬流过程是否完成,而自噬流的意思是自噬的完整动态过程,包括自噬体形成、自噬体与溶酶体融合及后续内含物的降解和回收。
-
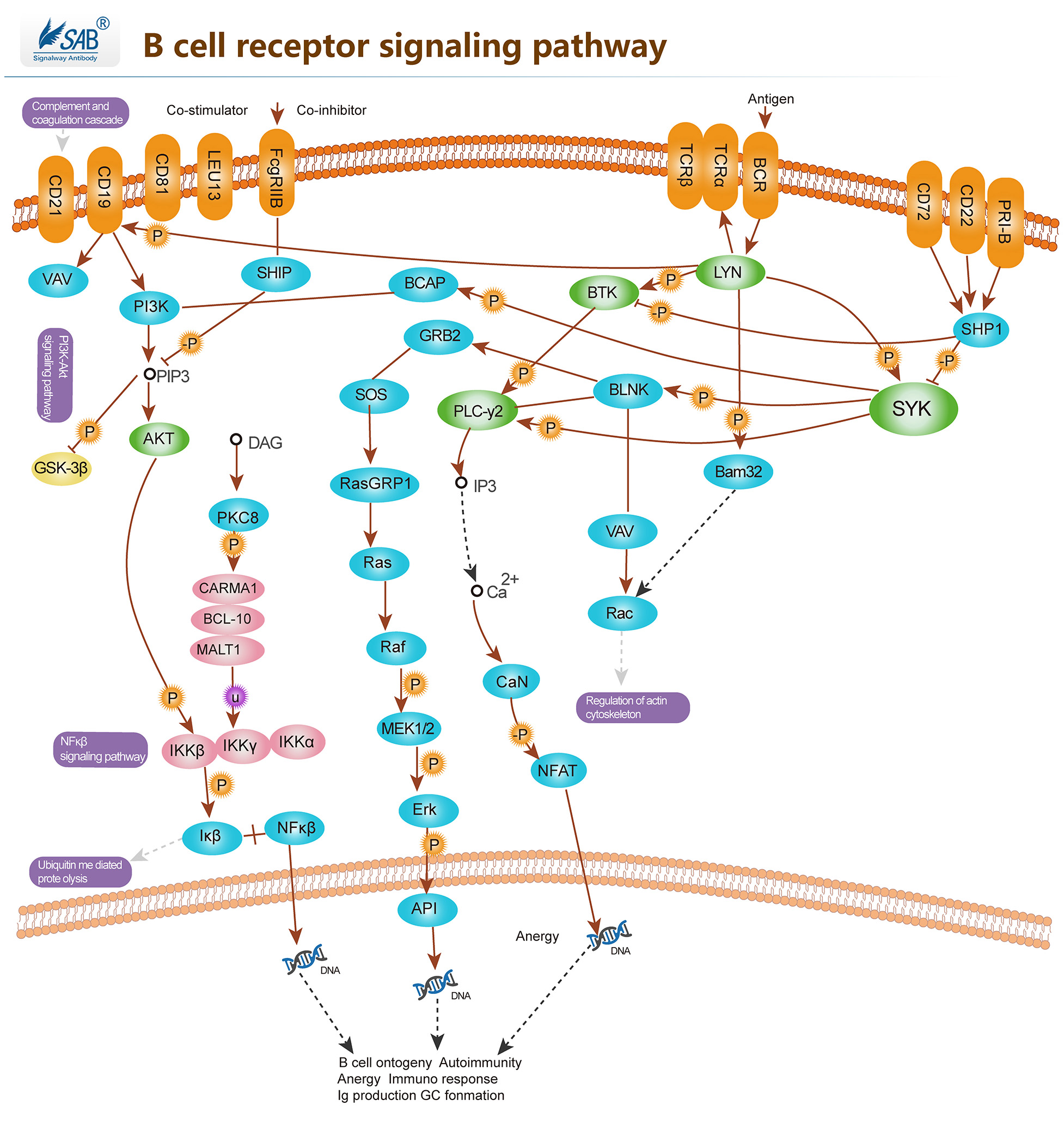
B CELL Pathway